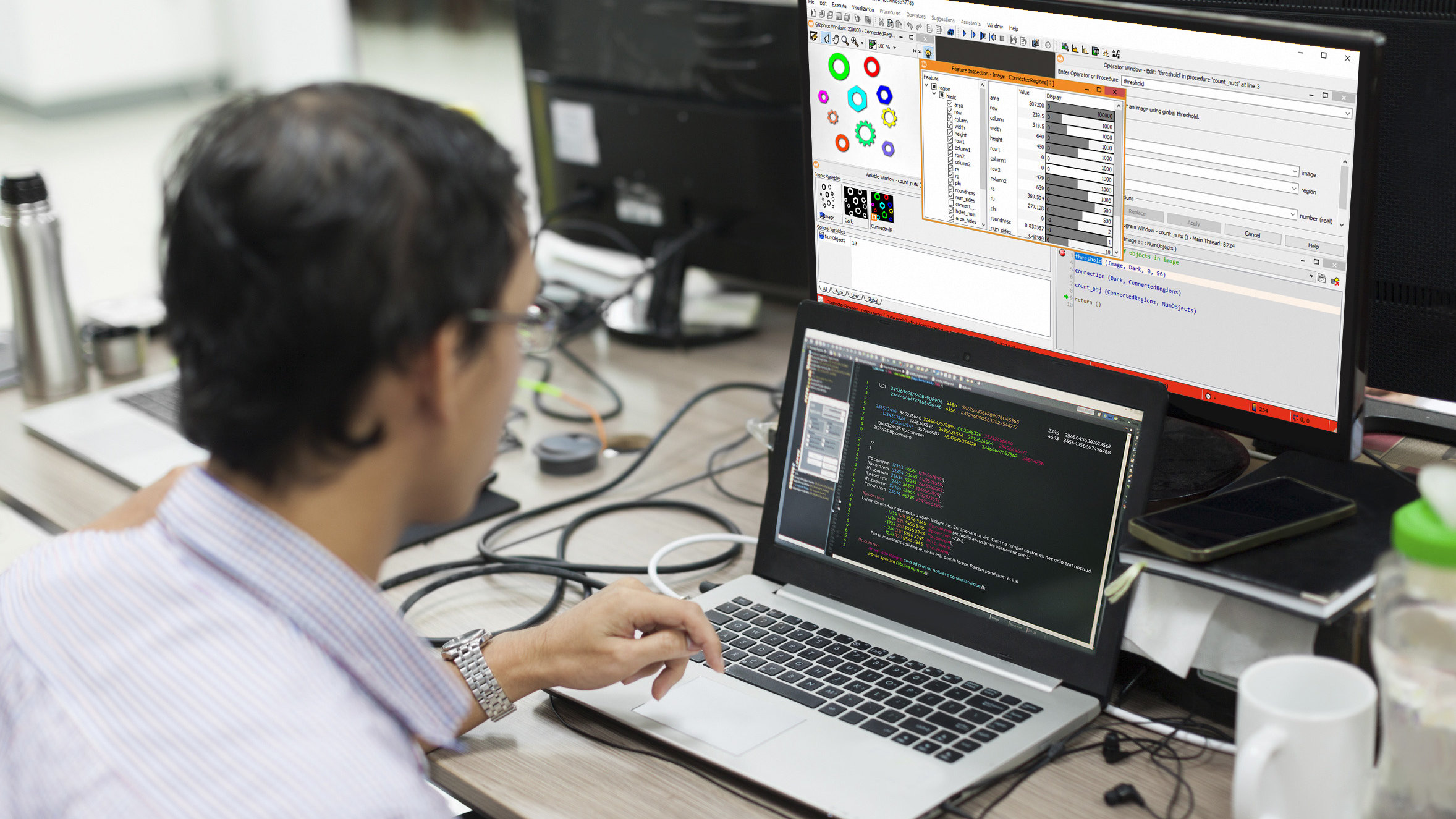
In this article, we aim to separate chickpeas and beans in the image below using image processing. We will use image segmentation and morphology algorithms for this task. Later in the article, we will review image morphology techniques, followed by step-by-step implementation in the powerful image processing software, HALCON, developed by a specialized image processing company in Germany. Additionally, in the video, we delve into more details and teach you the use of HALCON’s exceptional tools, the best image processing software.
For registration in the “Machine Vision Training Course with HALCON Software,” click here.
Download the Image Processing Program for Chickpea and Bean Separation in HALCON Software
To download the program for detecting and separating chickpeas and beans with HALCON software, enter your email to receive the download link. Once you download and install HALCON, you can run the program.
Morphology Techniques in Image Processing
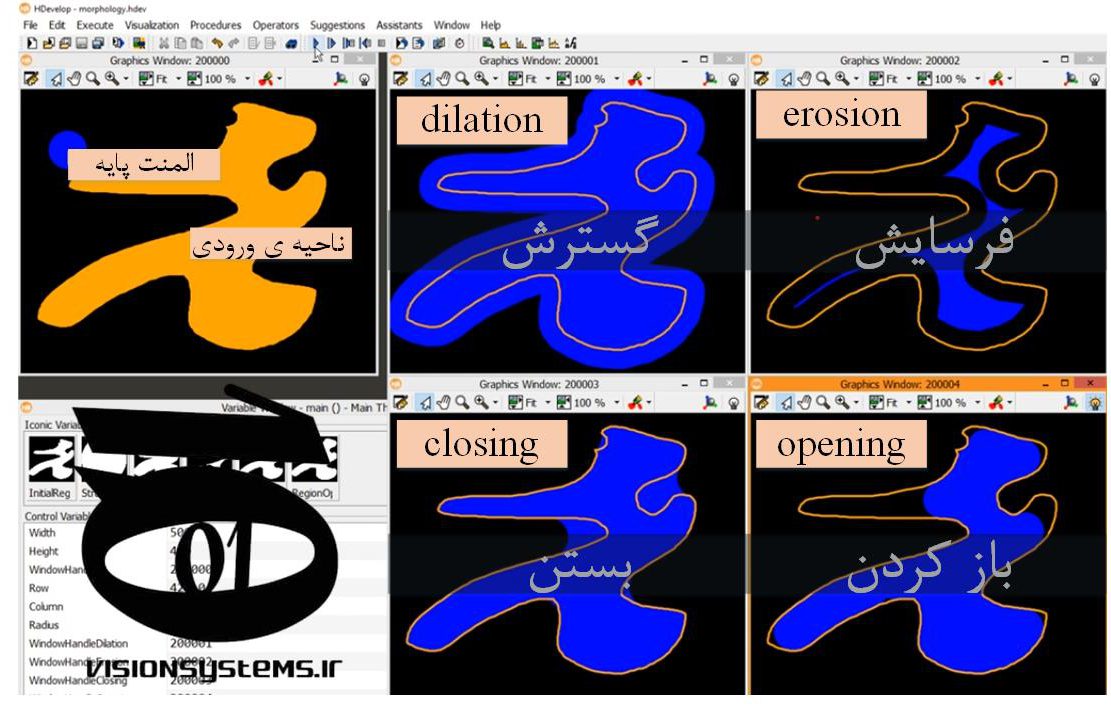
Morphology refers to the analysis and processing of geometric shapes. Four of the most important morphology operations are:
- Opening: Eliminates small shapes and protrusions from the image.
- Closing: Fills holes in regions.
- Dilation: Enlarges regions.
- Erosion: Erodes regions.
In the image below, we have outlined a sample region, and morphology operations are applied to it. The result of executing the four morphology operations is shown.
Coding Stages in Image Processing Program
To solve the problem of separating chickpeas from beans, there are three fundamental stages, each detailed in the following article.
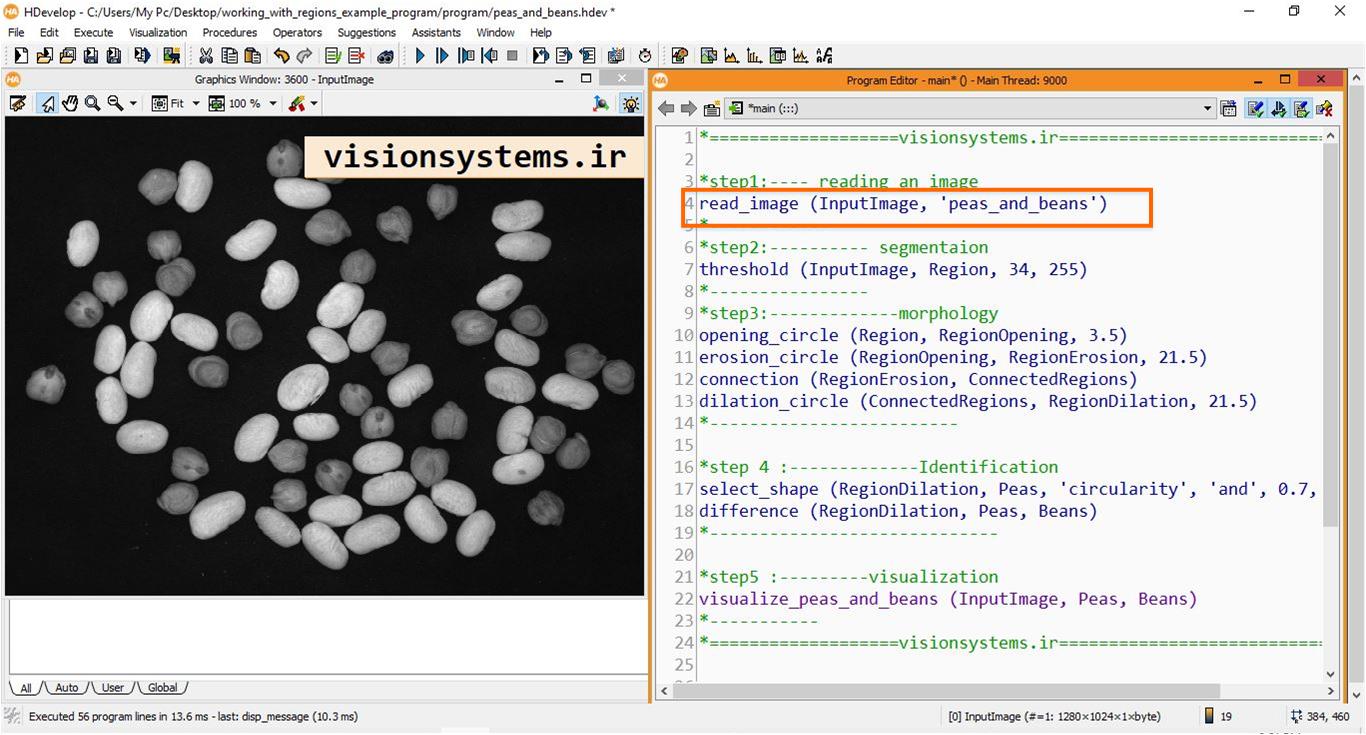
1- Reading the Image for Processing
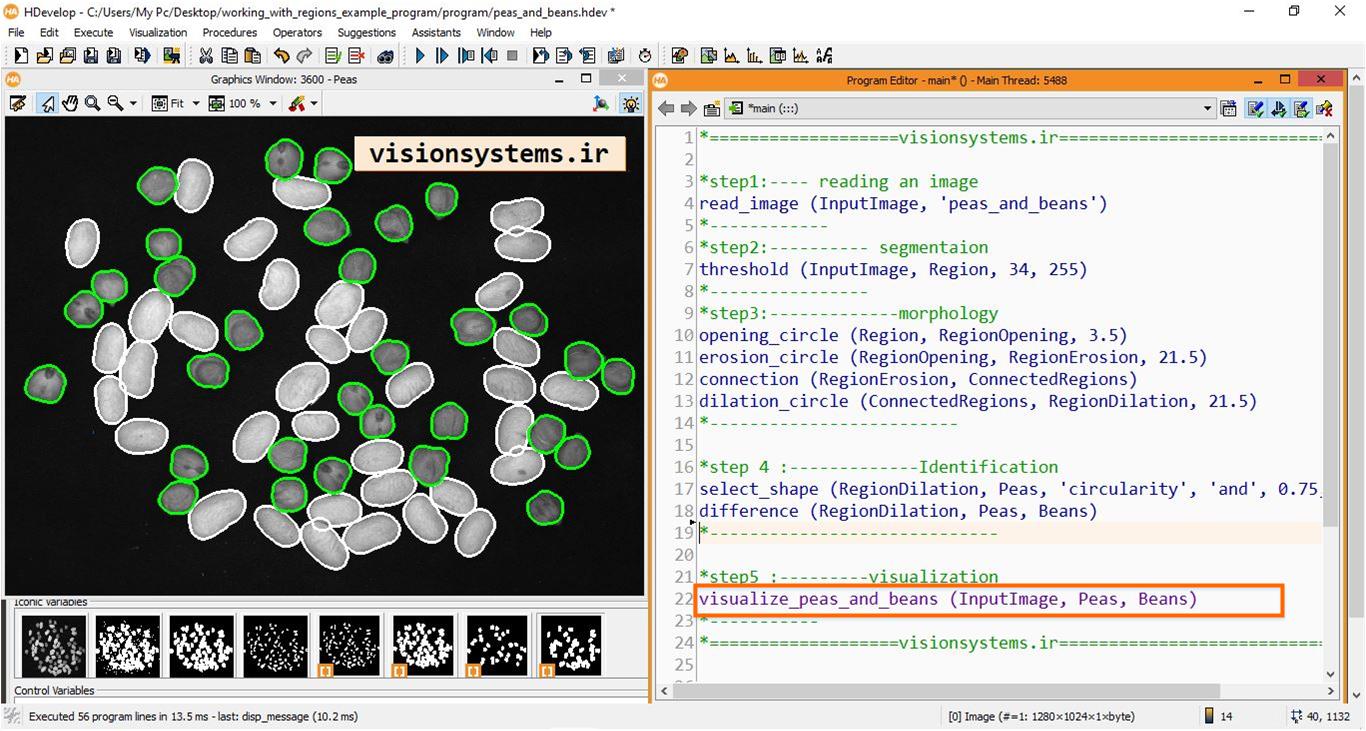
To read images in HALCON software, the read_image code is used. This code takes the name of the image provided next to the program (‘peas_and_beans’) as input. The result of running this code is that the image with the name InputImage is entered into the HALCON program.
read_image (InputImage, 'peas_and_beans')InputImage: Input image
2- Image Segmentation Using Morphology
Image segmentation aims to separate individual regions of chickpeas and beans. For this purpose, we need to perform the following five steps:
- Step 1: Remove the background by thresholding the image.
- Step 2: Remove noise using the opening technique.
- Step 3: Separate connected regions with erosion.
- Step 4: Segmentation of regions.
- Step 5: Use morphology to dilate the regions.
Each step will be explained in detail below.
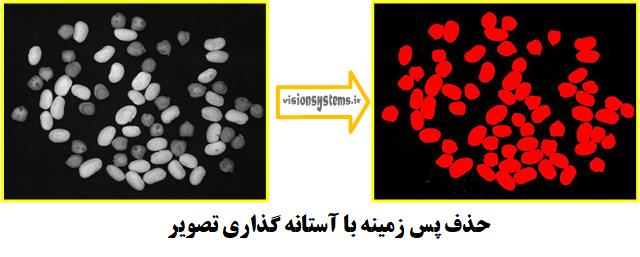
Step 1: Remove Background by Thresholding the Image
After reading the image, we need to separate chickpeas and beans from the image. For this purpose, we use thresholding on the image.
threshold (InputImage, Region, 34, 255)InputImage: Input image
Region: Region of chickpeas and beans
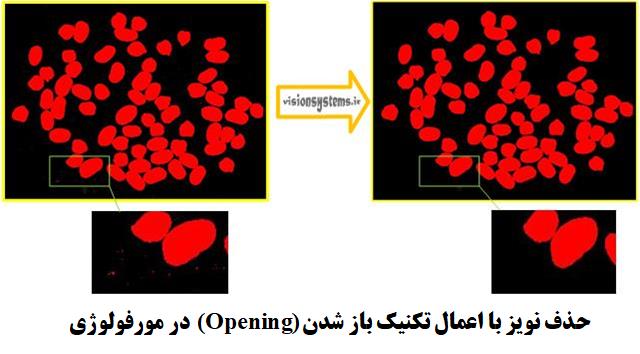
Step 2: Remove Noise with the Opening Technique
Using opening_circle, we can remove small regions in the image. In other words, this code takes the region obtained from the previous step (Region) as input, removes small regions, and places the result in the output (RegionOpening). The radius of 3.5 pixels is used in this code, effectively removing regions with a radius less than 3.5 pixels.
opening_circle (Region, RegionOpening, 3.5)Region: Input region
RegionOpening: Output region
Step 3: Separate Connected Regions with Erosion
To separate connected regions, we use the erosion_circle code. This code takes the region obtained from the previous step (RegionOpening) as input, erodes it with a circle of radius 21.5, and places it in the output variable (RegionErosion).
erosion_circle (RegionOpening, RegionErosion, 21.5)RegionOpening: Input region obtained from the previous step
RegionErosion: Region eroded with a circle of radius 21.5
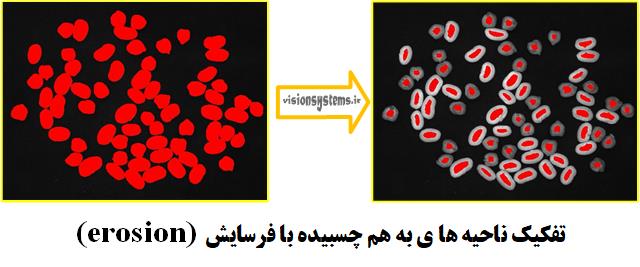
Step 4: Segmentation of Regions
So far, we have been able to separate the region related to chickpeas and beans from the background. Now, to access the region of each chickpea and bean, we need to break them down in the HALCON software. This is done using the connection code. This code takes the region obtained from the previous step (RegionErosion) as input and places the separated regions (ConnectedRegions) in the output.
connection (RegionErosion, ConnectedRegions)RegionErosion: Region obtained from the previous step (input)
ConnectedRegions: Separated regions (output)
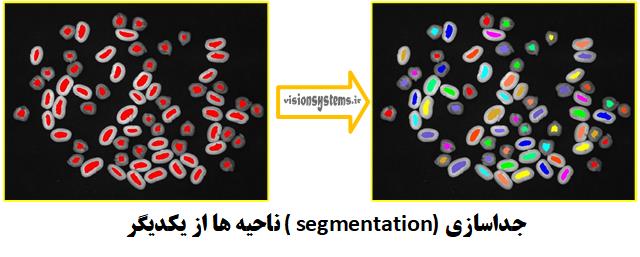
Using Morphology for Region Expansion (Dilation)
To accurately represent peas and beans, it is sufficient to expand the region obtained in the previous step using morphology. For this purpose, we use the code dilation_circle. This code takes the small regions obtained from the previous step (ConnectedRegions) as input, expands them with a circle of radius 21.5, and places the result in RegionDilation.
dilation_circle (ConnectedRegions, RegionDilation, 21.5)
ConnectedRegions: Regions obtained from the previous step (code input)
RegionDilation: Regions expanded with a circle of radius 21.5 (code output)
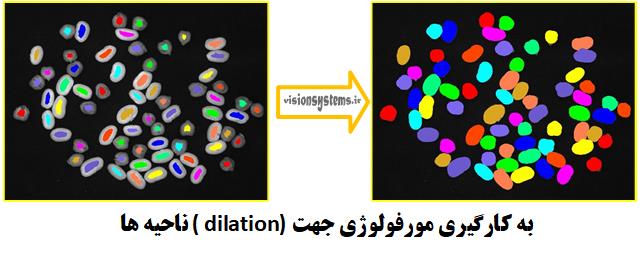
3- Detection and Separation of Beans and Peas Using Features
Detecting Peas
To detect peas from beans, we need to look for features that differentiate them. For this purpose, we use the circularity feature. This is done using the select_shape code. This code takes the regions of peas and beans as input and outputs those with circularity between 0.75 and 1 (which corresponds to the regions of peas).
select_shape (RegionDilation, Peas, ‘circularity’, ‘and’, 0.75, 1)
RegionDilation: Regions of peas and beans (code input)
Peas: Regions of peas (code output)
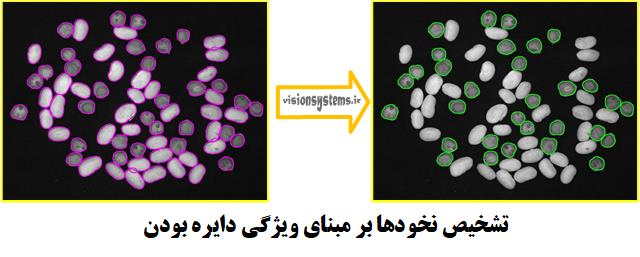
Detecting Beans
Now that we have detected peas, we can easily obtain beans. To do this, we simply subtract the regions of peas from the total regions. This is done using the difference code.
difference (RegionDilation, Peas, Beans)
RegionDilation: Entire region (sum of peas and beans regions)
Peas: Regions of peas
Beans: Regions of beans

Displaying the Results of Image Processing Program
To display beans and peas, the visualize_peas_and_beans function is used. This function takes the region corresponding to beans and peas along with the image as input and then displays them on the image. As you can see in the image, beans and peas are correctly separated from each other.
visualize_peas_and_beans (InputImage, Peas, Beans)
InputImage: Input image
Peas: Regions of peas
Beans: Regions of beans
You can consult with our experts for your projects. Consult with us.